History and status of aquatic breeding
The first aquaculture textbook was written in 475 BC (Fang Li, “Fish Farming Classic”), but little genetic improvement through breeding has been accomplished until recently. Even now, the vast majority of farmed fish are produced using low performing wild stocks (recurrently captured wild fry) or cultured stocks which often suffer from genetic inferiority due to erosion and inbreeding.
Only about 3 percent of world aquaculture production is based on broodstock originating from selective breeding programs. During recent decades however, substantial genetic improvement and subsequent increase in production efficiency has been well documented in some farmed fish species such as salmonids and tilapia. This improvement has been achieved through the implementation of quantitative genetic principles and adequate tagging systems.
Growth rate has been the first and predominant trait to be improved by selective breeding. A faster growing fish will increase the number of harvests and usually exploit the available feed resources more efficiently i.e. improved feed conversion ratio. However, in some aquatic species such as salmonids and in particular marine shrimps, there are tremendous losses in production due to infectious diseases. Therefore, there is also a definite demand for improved disease control systems through genetic improvement, pathogen screening, vaccines etc.
Although the aquaculture industry is developing rapidly, many operations are working under medium to small margins, with high investment and operating costs, and high risk due to inferior seed and disease problems. The increasing demand for aquatic proteins requires new solutions to increase production without depleting wild resources. It is beyond doubt that breeding will increase the overall aquaculture output and thus gain importance. Cost saving new breeding technology is being developed using molecular genetic methods to improve aquaculture seed at unprecedented rates.
Bioinformatics
New breeding technology can be applied to any species with a closed life cycle. Genomics, which may be defined as “methods to study the organization, structure and function of the genes of an organism,” provides a powerful tool to make quantum advances in aquaculture productivity. Well established breeding principles fortified with the latest genomics developments enable highly efficient genetic improvement in large scale breeding populations of main species groups like salmonids as well as in small breeding populations of niche-species.
Informatics is an additional technology required to manage and analyze the vast and rapidly accumulating amounts of molecular biology data generated through genomics.
The merger of molecular biology and informatics constitutes bioinformatics, widely held to be among the most rapidly developing fields of the 21st century; the others being information technology and telecommunication. Bioinformatics, really a true hybrid between molecular and computer sciences, changes the rules of both breeding and medicine by its novel approach to diagnostics, vaccinology, therapeutics, feed additives, drug discovery, gene therapy and genetic refinement/germplasm enhancement (superior seeds). Traditional classifications are less valid than before.
Normal genetic gain resulting from conventional selective breeding is 10 to 15 percent per generation. Growth rate has been the major trait subjected to successful breeding in salmon and tilapia. Studies have shown that the employment of genomic assisted genetic refinement strategies will increase this gain many fold, especially for traits which are hard or very expensive to record by conventional phenotypic tools. Such traits are, for example, disease resistance, filet quality (color), salt and temperature tolerance, and feed conversion ratio.
Some examples on how genomics can assist in speeding the genetic gain in breeding programs:
Genetic profiling
By DNA testing parents and offspring, one can with absolute security assign an individual to its parents. This is done by DNA analysis using so-called genetic markers such as microsatellites and Single Nucleotide Polymorphism (SNP). Such genetic profiling (also termed DNA fingerprinting) enables one to keep more individuals and more families in the breeding nucleus without need for separate tanks etc. and without confusing the paternity. This again provides the basis for higher selection intensity and higher genetic gain (about twofold the gain without DNA fingerprints, approximately 20 to 30 percent genetic gain in growth rate per generation).
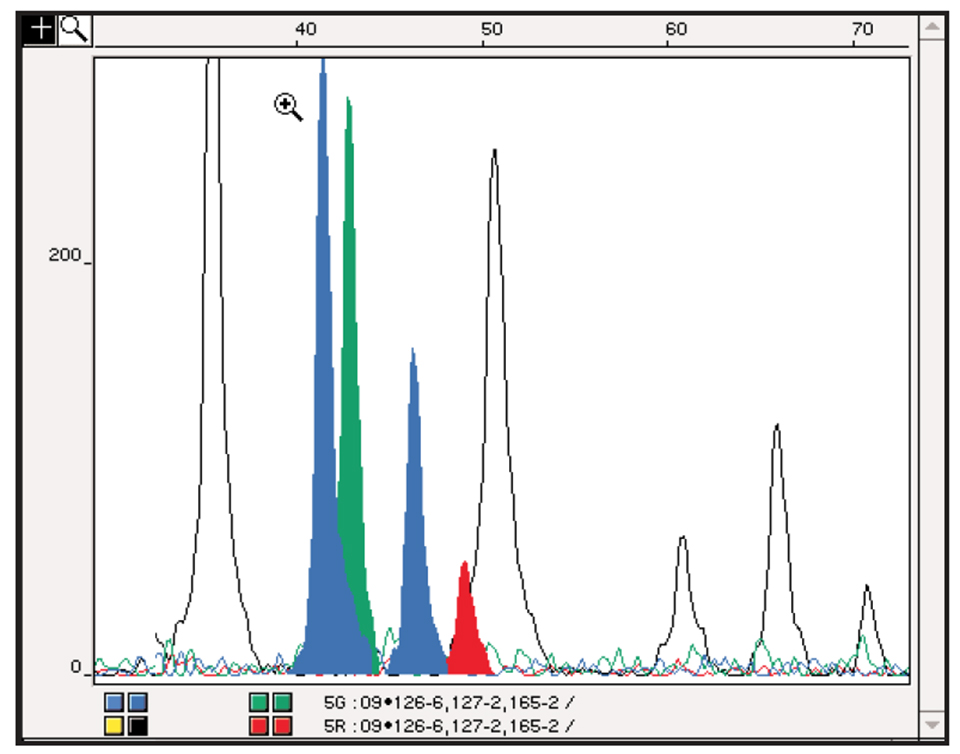
DNA fingerprinting is also a useful tool in stock identification for potential protection purposes, for characterizing genetic diversity in founder broodstocks and for wild stock management. DNA fingerprinting systems have been developed for several aquaculture species and such systems are ready for salmonids, tilapia, sea bass and others. An example of a DNA profiling analysis is shown in Fig. 1.
Exploiting information from economically important genes
The active use of information about identified genes controlling economically important traits such as growth rate, meat and skin color, salt water and disease resistance, filet yield and quality, provides another quantum leap in genetic refinement. Such information can be utilized by employing transgenic strategies which involve modifying the genetic make up of a strain or a species by transferring DNAor genes into an individual at an early stage (fertilized egg) with artificial methods. Although some examples of transgenic organisms have been seen at the research stage in aquaculture species, there is probably substantial distance from experimentation to commercial success and general acceptance. Some major reasons are: technical hurdles in terms of stability and side effects, market/community resistance together with regulatory restrictions or rejections in many countries.
Another way of utilising such gene information is through marker assisted selection (MAS), where the broodstock is DNA tested in order to reveal if the individuals possess superior or inferior gene variants for the specific trait. Hence, the breeding value can be predicted at an early stage (fertilized egg or fry level) and new traits with new market advantage can be selected in a cost efficient way through MAS. Several genes controlling important production traits have been identified in farm animals and such genes are also starting to be revealed in aquaculture species as well. A gene scan has identified genomic regions in tilapia associated with growth and body shape, the latter influencing filet yield. Mapping of those traits is being refined and the opportunity now exists for “hunting” of genes controlling sex, filet coloration in salmon and salt water and temperature tolerance in tilapia.
DNA-chips
The above enhancement procedures require cost efficient DNA typing systems with high throughput capacity. An emerging technology platform with impressive and diverse development both at the lab bench and at the market level is the bio chip or DNA chip. It enables breeders to DNA test an extremely large number of individuals for a very high number of genes and markers at speeds far beyond conventional methods. This greatly facilitates marker assisted selection or DNA fingerprinting. DNA chips are small glass slides (usually not larger than 2 square centimeters) where several thousands of different DNA analyses can take place simultaneously. Such DNA chips are now invading the human scientific community as well as the farm animal science and services. DNA chip techniques are being developed for DNA fingerprinting and pathogen screening in several aquaculture species. At a later stage chips for economical important genes will be developed as well.
Studies show a three-fold and a 13-fold genetic gain for growth rate and feed conversion ratio, respectively, compared to the gain of conventional phenotype breeding when the above strategies are implemented. Novel breeding software and breeding designs utilizing DNA profiling and later marker assisted selection are now available for any species.
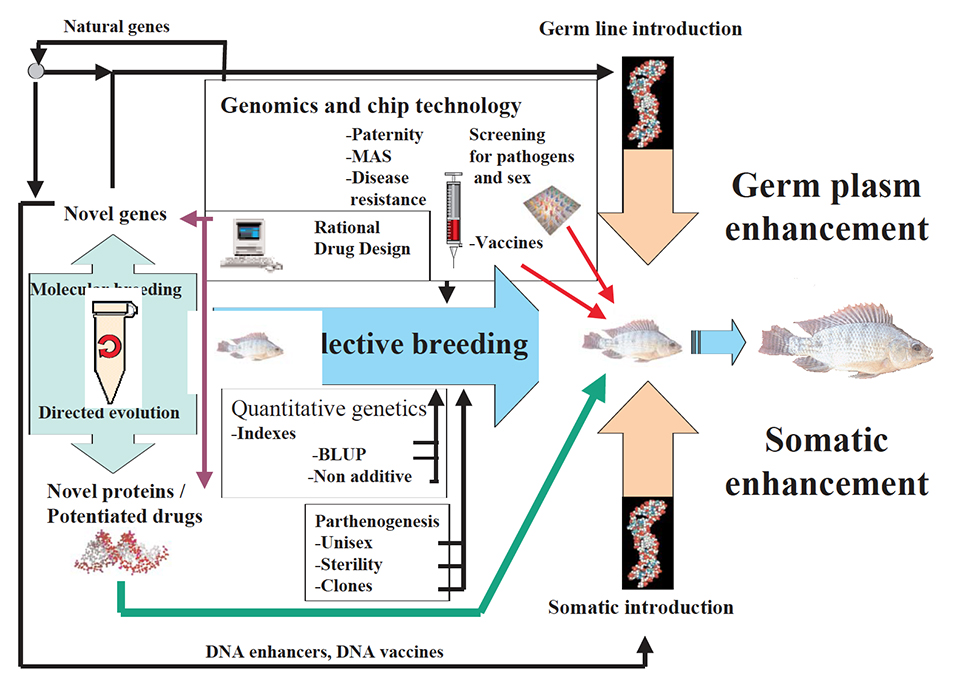
Genetic improvement can be accelerated with genomics and bioinformatics tools with various “delivery systems” like marker-assisted selection, somatic enhancement systems (stimulatory products and vaccines resulting from natural or novel genes and proteins), pathogen screening and germ line insertions (Fig. 2). Several of these strategies are still not realistic due to technical, safety, ethical and market perception reasons. GenoMar is involved with bioinformatics enhanced breeding, pathogen screening and vaccine prototypes, which involve no gene modification of the fish.
Molecular disease control systems
Genomics tools are also successfully employed in the detection of fish pathogens and in the development of new vaccine generations, so-called DNA vaccines. Through powerful DNA techniques (e.g. PCR technology), one can detect the presence of infectious agents in the tissue of individuals as well as in the environment at very high levels of specificity and sensitivity. This is now a routine technique in human and farm animal diagnostic services, and it is also under establishment in the aquaculture industry. A DNAbased diagnostic test for Infectious Salmon Anaemia (ISA) has been developed and similar tests for other pathogens are under development. Eventually pathogen screening will be run on chip platforms as well.
Several research groups are now involved in developing DNA vaccines against pathogens where traditional vaccines do not exist or show little or no effect. Such vaccines are based on the injection into somatic cells of the fish of small, harmless DNA pieces (constructs) resembling genes from a pathogenic organism and such eliciting a protecting immune response against the same pathogen. DNA vaccines are under development against various fish pathogens. Conclusion Bioinformatics is a new state-of-the-art technology which offers greater levels of genetic enhancement and disease control, which will improve the profitability and sustainability of aquaculture in the third Millenium.
(Editor’s Note: This article was originally published in the December 1999 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Øystein Lie
GenoMar AS (Formerly BioSoft AS)
Oslo Research Park
Gaustadalleen 21
N-0349 Oslo, Norway
Website: www.genomar.com -
Aaudun Slettan
GenoMar AS (Formerly BioSoft AS)
Oslo Research Park
Gaustadalleen 21
N-0349 Oslo, Norway
Website: www.genomar.com -
Hans Magnus Gjøen
GenoMar AS (Formerly BioSoft AS)
Oslo Research Park
Gaustadalleen 21
N-0349 Oslo, Norway
Website: www.genomar.com
Tagged With
Related Posts

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Aquafeeds
Breaking the 20 percent soy barrier in fish feed
Reduced performance in fish fed high-soy feeds has been blamed on antinutrients, low methionine content and palatability issues. Pretreatment to inactivate anti-nutritional compounds and supplementation with amino acids improves soy-based feed performance, but not to control levels.

Health & Welfare
Developmental transcriptomes from penaeid shrimp
To develop an alternative mechanism for genetic copyright of improved shrimp lines, the authors used a bioinformatics approach to identify germ line genes that could be potentially targeted to ablate the germ line.
Health & Welfare
Nutrition and genes research at Virginia Tech
Transcriptomics studies highlight impacts of different treatments on specific genes that can be targeted via nutritional intervention to increase farmed animal performance or welfare.



