Molecular markers can detect differences in natural and farmed tilapia populations

The last two decades witnessed the development of genetically improved Nile tilapia stocks in Asia, particularly the Philippines. Major genetic improvement programs introduced new “strains” that are now raised at many commercial farms worldwide.
Because locally available stocks were commonly believed to be genetically inferior, improved breeds such as the GIFT, GMT, and FaST tilapia sold well when first introduced commercially. Most entrepreneurial farmers are prepared to pay a premium for such tilapia seed in the hope of raising their returns through shorter culture periods or lower food costs.
Genetic screening
Genetics in tilapia aquaculture has mostly involved modification through conventional selection (mass selection, family selection, hybridization, and other means) and modern manipulation methods such as transgenesis and chromosome set manipulation. Another genetic application that is becoming an integral part of tilapia culture and broodstock management is the use of biochemical or molecular markers.
With the advent of polymerase chain reaction technology, molecular markers have been developed to detect differences in natural and farmed tilapia populations. Mitochondrial DNA restriction fragment length polymorphism mtDNA-RFLP), amplified fragment length polymorphism, multilocus fingerprinting, and randomly amplified polymorphic DNA are some of the techniques used to delineate stock differences. With the development of the tilapia genome map at the University of New Hampshire, the genetic structure of wild and farmed Nile tilapia stocks is now readily analyzed through highly variable microsatellite DNA markers (msDNA).
The GIFT project was the only major tilapia-breeding program in the Philippines that investigated the genetic variability of its founder stocks (four African and four Asian Nile tilapia stocks) using allozymes, not the more versatile modern molecular markers. These markers could determine the genetic impacts of the various improvement approaches, and also identify and perhaps protect the integrity of the improved stocks.
Characterization using DNA markers
Since molecular markers can resolve high levels of polymorphism, they are useful in describing genetic variation among stocks. With the potential of using markers as tools to effectively assess, manage, and conserve the genetic diversity of farmed tilapia, the genetic structure of improved and domesticated Nile tilapia stocks in the Philippines was determined in a collaborative study between SEAFDEC and the Laboratory of Applied Population Genetics of Tohoku University.
Two marker systems, msDNA and mtDNA-RFLP, were used to evaluate domesticated NIFI and Israel tilapia, and GIFT, GMT, FAsT, and SEAFDEC selected Nile tilapia stocks (Table 1). Thirty fin clip samples taken randomly from each of the stocks were collected and preserved in ethanol for analysis in Japan.
Eguia, Details of the farmed Asian stocks of Nile tilapia, Table 1
| Stock | Generation | Remarks |
|---|---|---|
| NIFI | F2 | Obtained in 1987 from the National Inland Fisheries Institute in Bangkok, Thailand, where it is known as Chitralada. |
| Israel | F2 | Acquired in 1979 by the Philippine Bureau of Fisheries and Aquatic Resources from the Gan Shmuel Hatchery in Israel. Disseminated to local farmers from 1970s to mid-1980s. |
| FaST | S11 | Obtained from the Freshwater Aquaculture Center of the Central Luzon State University in 1993. From a within-family selection program. Founders consisted of Israel, Singapore, and Taiwan stocks. |
| SEAFDEC-selected | S11 or F11 | Originally selected from the second-generation offspring of NIFI, (S4) is the fourth selected generation in a size specific mass-selection program. |
| GMT | F1 | Genetically male tilapia (GMT) maintained by PhilFishgen (Nueva Ecija, Philippines). The founder stock used to develop the YY-supermale tilapia in GMT production was obtained from Lake Manzala, Egypt, by the University of Wales, Swansea, through the University of Stirling. This batch is a cross between an F4 selected line (F) x Egypt-Swansea YY (M). |
| GIFT G9 | G9 | Ninth-generation genetically improved farmed tilapia (GIFT) produced by crossing best-performing strains from Africa and Asia using combined family selection. Maintained by GIFT Foundation International Inc., Nueva Ecija, Philippines. |
Genetic diversity
Genetic diversity based on a five-microsatellite loci multiplex was more informative in characterizing stock differences than mtDNA-RFLP markers that were based on only 14 restriction morphs. Variability rankings among the Nile tilapia stocks revealed some inconsistencies between the marker systems results.
Contemporary msDNA data showed that GIFT tilapia had the highest mean expected heterozygosity (Hexp = 0.813) and the greatest allelic diversity (mean number of alleles per locus, A = 10), while GMT had the lowest (Hexp = 0.666, A = 6.4).
The improved breeds, especially GIFT and FaST (Hexp = 0.788), were generally more diverse than the unselected NIFI and Israel stocks, reflecting the fact that they were derived from synthetic mixed-base population stocks and have been managed to minimize inbreeding. In contrast, mtDNA haplotype frequencies showed that the unselected stocks NIFI (haplotype diversity, h = 0.805) and Israel (h = 0.648) were the more variable stocks.
Although the mean genetic variability based on msDNA analysis was moderately high in all the tilapia except the NIFI, the stocks exhibited remarkably significant homozygote excess, indicating some degree of inbreeding. In addition, the greater variation in the total genetic diversity of the surveyed stocks was explained principally by differences at the individual level (99.61 percent based on msDNA analysis and 61 percent based on mtDNA results) and not the stock level.
Results also revealed some incongruities. MtDNA nucleotide divergence values showed the NIFI and SEAFDEC-selected animals as distantly related, while msDNA data indicated genetic similarity between the two stocks. NIFI was the founder stock used in the development of the SEAFDEC-selected line, so the relationships indicated by msDNA analysis appeared more reliable in this context.
Stock management
These results have implications for the conservation and management of farmed tilapia stocks. While the study would have benefited from the use of known inbred and outbred “reference” stocks, both molecular marker methods indicated some inbreeding in the majority of the farmed Asian Nile tilapia stocks.
Inbreeding is a product of small founder stocks and/or poor broodstock management. Knowledge of the extent of inbreeding in farmed tilapia stocks through DNA markers promotes an increased awareness of the importance of proper broodstock management in minimizing the negative consequences of inbreeding depression.
The tilapia test stocks were less differentiated than might have been thought, as seen in the very low percentage of molecular variation due to stock differences. This may have been due to the dominance of Egyptian founding stocks in several of the Asian strains. It is possible the marker frequencies noted in the surveyed stocks tended toward common alleles and haplotypes. In any case, the fact that the synthetic GIFT strain had the highest genetic variability and allelic diversity meant that the inclusion of “new” alleles from the African germplasm generated a sustained increase in GIFT’s genetic variability relative to those derived only from Asian founder stocks.
Finally, since the genetic variation in the surveyed Asian stocks was due mainly to individual-level differences, selective-breeding methods that exploit individual-level genetic variation should be given more emphasis in future genetic improvement programs.
Genetic monitoring
Apart from characterizing stocks, DNA markers can be used to monitor genetic changes in selected and domesticated stocks. The study above used genetic markers to assess the effects of a simple two-step size-specific mass selection on the genetic variability of the NIFI stock.
Fin clips from individuals of two generations of an unselected control stock (first C1 and fourth-generation C4) and three generations of the SEAFDEC-selected Nile tilapia (S1, S2, and S4) were analyzed for msDNA variation at the same five loci. Apart from genetic variability indices, growth, response to selection, and inbreeding coefficients at each generation of selection were estimated.
Microsatellite analyses showed a reduction in some genetic variability indices (A and mean Hexp) in both the selected and control stocks after four generations. Mean Hexp levels were reduced significantly, while an apparent though nonsignificant reduction in the number of alleles was noted in S4. The decrease in allelic diversity was due to the elimination of low-frequency alleles in the course of selection, while the decrease in genetic variability levels implied increased homozygosity or inbreeding in the successive selected generations.
This happened as fewer and genetically less diverse individuals contributed as effective parents to the next breeding generation. This inbreeding appeared to have a negative impact on the growth advantage of the selected stocks over the control stocks, as mean growth rate differences were reduced in the later generations. The control stocks also showed some reduction in allelic diversity and genetic variability, brought about by unintentional stabilizing selection and largely random genetic drift, which occurs naturally in limited breeding populations such as aquacultured stocks.
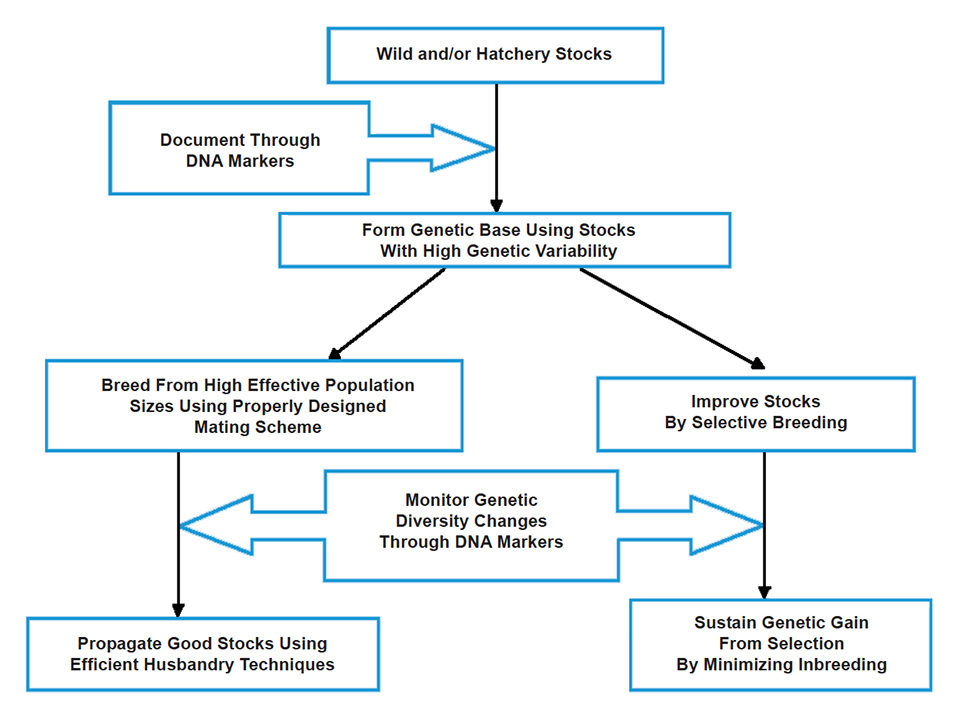
Maintain diversity
With genetic marker data, appropriate broodstock management and genetic improvement methods can be formulated to protect the diversity of existing tilapia genetic resources. Thus, whenever possible, DNA markers should be integrated in schemes that promote the sustainable management of farmed tilapia (Fig. 1). Sustainable management can be achieved by both developing tilapia lines that can be utilized immediately for farming and maintaining highly genetically variable populations as valuable genetic resources for future use.
Baseline data on the genetic diversity of farmed tilapia stocks provides good reference for aquaculturists. Ironically, before selected tilapia stocks were developed, wild and farmed tilapia populations were poorly managed. Aquaculturists were partly blamed for genetic stock deterioration because of indiscriminate stock transfers and the application of inappropriate broodstock management practices. With the availability of advanced genetic-monitoring techniques and increased awareness in genetic resource management, aquaculturists now have the means to protect the diversity of farmed tilapia stocks.
(Editor’s Note: This article was originally published in the July/August 2006 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Maria Rowena R. Eguia, Ph.D.
SEAFDEC Aquaculture Department
Binangonan Freshwater Station, Tapao Point
Binangonan 1940 Rizal, Philippines[104,112,46,103,114,111,46,99,101,100,102,97,101,115,46,100,113,97,64,97,105,117,103,101,114,109]
-
Prof. Nobuhiko Taniguchi
Laboratory of Applied Population Genetics
Tohoku University
Sendai, Japan
Tagged With
Related Posts

Intelligence
Aquaculture UK: Stepping up to the plate
There’s considerable opportunity to grow the UK aquaculture industry. At the Aquaculture UK exhibition and conference in Aviemore, Scotland shows the way.

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Health & Welfare
Candidate gene markers for selective breeding of CyHV-3-resistant carp, koi
Common carp and koi producers around the world have suffered substantial financial losses due to a contagious disease caused by cyprinid herpesvirus-3.

Responsibility
Changing the production chain of Amazonian pirarucu
Studies in Brazil over the last five years addressed the challenges facing commercial producers of the important, air-breathing, freshwater fish pirarucu in order to support and expand production.


