Marine Biological Laboratory identifying internal factors that control the growth and development
In the mid-1960s, scientists first identified the bay scallop (Argopecten irradians) as a good candidate for aquaculture and established many of the techniques behind the current culture methodology. Current technology is primarily focused on getting quality, market-sized scallops to consumers in reduced time with minimum financial expenditure.
Scallop culture
Scallops have a very high growth rate, reaching sexual maturity in about six months. Bay scallops reach market size in one or two years in the wild or significantly less time under controlled conditions. Because bay scallops are mobile, grow-out usually occurs in suspended nets, as opposed to the bottom culture employed for oysters and clams.
The single large adductor muscle that evolved in bay scallops acts to open and close the shell with great force. This muscle is the most prominent feature observed when the shell is removed, and also the primary portion of the scallop desired by seafood consumers, particularly in the United States. As with any animal muscle, scallop adductor muscles are rich in protein and glycogen.
Great taste and high nutritional value have put the muscles of scallops in high demand for many years. When bay scallops are available, their market value is extremely high. For example, in the New England region of the United States, local bay scallops can sell for U.S. $35 to 48 per kilogram in the retail market.
Control of development
A primary problem with bay scallop culture in the United States is the animals’ high rate of mortality during early development and overwintering. One focus of the research at the Marine Biological Laboratory in Massachusetts, USA, is to identify the internal factors that control the growth and development of bay scallops.
By understanding what specific factors are involved, scallop larvae could be stimulated to set and grow faster, potentially decreasing mortality rates and the time needed to get the scallops to market size. Additionally, as factors important for growth are identified, a selective-breeding process could be initiated to select for scallops with desired traits.
Understanding genetic factors
In research supported by the U.S. Department of Agriculture, multiple approaches are being taken at the Marine Biological Laboratory to better understand the genetic factors involved in the growth and development of bay scallops. Specific projects include an expressed sequence tag project and the use of a polymerase chain reaction-based, differentially expressed gene technology.
Expressed sequence tags are small fragments of DNA sequence generated by single-pass sequencing. Sequencing bits of DNA that represent genes expressed in specific tissues or during different stages of development provides researchers with information necessary to help identify unknown genes and map their position in the genome.
EST analysis
For bay scallops, expressed sequence tag analysis is performed on four cDNA libraries generated from bay scallop larvae taken at three developmental stages and one adductor muscle library. A summary of the sequences obtained from adductor muscle tissue is shown in Table 1.
Roberts, Expressed sequence tags from a bay scallop adductor muscle, Table 1
| Sequence | Percentage |
|---|---|
| Cell structure | 33% |
| Metabolism | 31% |
| Unclassified | 11% |
| Cell signaling | 8% |
| Binding proteins | 7% |
| Immune-related | 7% |
| Gene/protein expression | 3% |
To date, over 2,000 sequences from the bay scallop are available to the public as part of the National Center for Biotechnology Information’s database. Several of the genes have been identified based on sequence homology. The next step in the research is to begin to fully characterize some of the more promising genes to understand their function in bay scallop growth and locate their position in the genome.
Differentially expressed genes
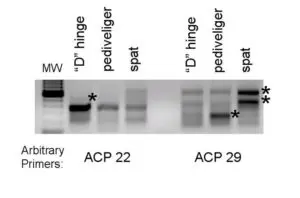
(indicated by asterisks) during bay scallop larval development. The two-stage polymerase chain reaction was performed using two arbitrary primers.
The second, more directed molecular approach being taken to understand the internal factors controlling development is the isolation of differentially expressed genes. These genes are all expressed differentially in the mRNA levels of different samples. To accomplish this objective, developing bay scallops were taken prior to (“D” hinge), during (pediveliger), and following metamorphosis (spat).
After RNA isolation, a commercial kit was used to amplify via polymerase chain reaction the differentially expressed genes. An example of a gel with the genes visible is shown in Fig. 1.
The premise behind this approach is that a relevant trait is associated with the up or down regulation of a gene. Currently, the identity of the genes in Fig. 1 is not known, but additional studies will help researchers understand how the genes are involved in development.
(Editor’s Note: This article was originally published in the June 2004 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Steven Roberts, Ph.D.
Program in Scientific Aquaculture
Marine Biological Laboratory
7 MBL Street
Woods Hole, Massachusetts 02543 USA[117,100,101,46,108,98,109,64,115,116,114,101,98,111,114,115]
Tagged With
Related Posts

Intelligence
Behold the nutritious oyster
Oysters provide important, natural filtration of water and are an important component of many healthy coastal ecosystems because their active filtering can help improve and maintain water quality. For many coastal communities, oysters are an important food resource and excellent sources of protein and amino acids, zinc, selenium, iron and B-vitamins.

Health & Welfare
Formulated diets for bivalve spat replace algae in hatchery trials
Although the hatchery rearing of bivalves currently relies heavily on the mass production of microalgae, new feed alternatives are being developed.

Innovation & Investment
Maine scallop farmers get the hang of Japanese technique
Thanks in part to a unique “sister state” relationship that Maine shares with Aomori Prefecture, a scallop farming technique and related equipment developed in Japan are headed to the United States. Using the equipment could save growers time and money and could signal the birth of a new industry.

Innovation & Investment
Making algae can get expensive. Innovations aim to bring costs down.
Microalgae play a key nutritional role in larval shellfish culture. Photobioreactors may be a simplified, cost-effective form of algae production.


