Fast, easy, reliable method offers great potential for all aquaculture systems

There is a widespread need for cultivation-free methods to quantify the viability of microbial communities in aquatic environments. This demand also exists within the aquaculture industry, where rapid and reliable methods for measuring bacterial activity and bioavailable organic matter in water are increasingly required.
Microbial water quality assessment has the potential to provide insights into the temporal and spatial dynamics of bacterial communities within aquaculture systems as a supplement to general chemical water quality parameters. This is of particular relevance to recirculating aquaculture systems (RAS) that are characterized by a continuous input of bioavailable substrates, high nutrient levels and long retention times all favoring heterotrophic bacterial growth.
The use of methods for assessing the bacterial and the broader microbial activity in water may contribute to gain additional knowledge of potential effects and interaction of various factors (i.e. feed composition, feed loading, hydraulics and mechanical, biological and chemical treatments) on bacteriology and growth potential in RAS. Easy and reliable methods are therefore crucial to the ongoing development of RAS because they can provide data for baseline conditions as well as detection of sudden unforeseen deviations.
Determination of biological oxygen demand over five days (BOD5) is a common method to quantify the bioavailable organic matter in aquaculture water. The method is informative on biodegradable organic matter but is inexpedient with respect to time. Traditional methods, such as plate counting to obtain colonies forming units (CFU) are sometimes used to predict changes in relative numbers of bacteria in aquaculture water samples.
This article – adapted and summarized from the original publication in Aquacultural Engineering – describes our proposed, rapid, simple, inexpensive and reproducible method that reflects the microbial enzymatic activity of both free living and particle associated bacteria, as well as potential contributions from other microbiota. The underlying principle takes advantage of H2O2 decomposition (also referred to as degradation, elimination, reduction or decay) which is primarily a biological process and hence related to the microbial composition of the water.
Our study was partly funded by the COFASP ERA-NET partners, which has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 321553 and the Danish EPA Pesticide Research Programme (j. no. 667-00199). We appreciate the analytical support from Brian Moller and Ulla Sproegel and the management of experimental RAS by Ole M. Larsen and Rasmus F. Nielsen from DTU Aqua Section for Aquaculture, Hirtshals.

Study setup
The method of this assay is based on adding a well-defined quantity of H2O2 to a raw water sample and quantifying the decomposition of H2O2 over time under controlled conditions. Under constant conditions (nominal H2O2 concentration, temperature and mixing), the decomposition rate reflects amounts of total enzymatic activity of bacteria and any eukaryotic microbiota present (planktonic and particle bound).
For all batch experiments, the H2O2 and chemical oxygen demand (COD) concentrations as well as the 5-day biological oxygen demand (BOD5) were measured in at least duplicates and the average values were used. Water volumes, H2O2 concentration and sampling frequency are modifiable as long as consistent mixing and temperature control are provided. An applied fixation reagent terminates the H2O2decomposition and forms a stable color complex allowing flexible sampling and subsequent spectrophotometric analysis.
The water samples used were all collected from a pilot scale freshwater recirculating aquaculture system (RAS) operated with rainbow trout at different densities and feed loadings. To investigate the effects of filtration on H2O2 decomposition kinetics, water was collected from a RAS operated at constant conditions for a period of four months. To compare H2O2 decomposition rate constants with BOD5analysis, water samples were collected from six different RAS over a three-week period). Finally, H2O2 decomposition rate constants were compared with micro particle numbers based on water samples from 12 different RAS operated at constant conditions.
For additional information on general description of the study; the procedure for H2O2 decomposition assay; reagents and chemical methods used; theory and calculations; description of RAS systems for samples; and statistical analyses, please consult the original publication or the first author.

Results and discussion
A linear standard curve based on H2O2 standard solutions from 0.5 to 25 mg H2O2/L was used to measure H2O2 in water samples spiked with H2O2. The H2O2 concentration was then calculated as [H2O2] = (Abs432 −0.0022)/0.0132 with that particular reagent. The level of detection of the applied method was 0.16 mg/L H2O2, corresponding to ≤0.002 Abs units, and H2O2 concentrations were adjusted for background absorbance and used to calculate decomposition rate constants for H2O2.
Regarding the effects of filtration, the H2O2 decomposition rate constants derived from raw and pre-filtered RAS water confirmed that the majority of H2O2 decomposition was related to particle associated bacterial activity. The rate constant in 0.2 μm sterile filtered RAS water at 22 degrees-C was 0.038/hours with a corresponding H2O2 half-life of T½ =18.2 hours. Planktonic bacteria and small aggregates in the size range from 0.2 to 1.6 μm had a seven-fold higher decomposition rate constant with k=0.263 h−1 (T½=2.6 hours), while the H2O2decomposition rate constant in unfiltered RAS water at 22 degrees-C was 1.425 −1(T½=0.49 hours).
The COD content of the unfiltered RAS water samples was 88.5 ± 0.1 mg O2/L, while 1.6 μm and 0.2 μm filtration reduced the COD content to 40.1 ± 0.2 mg O2/L and 35.9 ± 0.2 mg O2/L, respectively. These values, along with accumulated nitrate and phosphate, indicates that RAS are nutrient rich environments with both dissolved and particulate organic matter favoring heterotrophic bacterial growth as opposed to the oligotrophic conditions in some natural waters. The direct correlation between biodegradable organic matter and rate of H2O2 decomposition was recently confirmed in RAS with different feed loadings, in RAS with acetate supplements and in a survey of seven commercial Danish model trout farms.
The abundance and distribution of planktonic bacteria versus particle associated bacteria and small eukaryotic microorganisms are likely to differ among individual RAS and over time. The exact causes, mechanisms and potential implications require further research. Site specific conditions and associated microbial composition may also reveal interesting enzymatic contributions from bacteria, algae and protozoa.
The H2O2 decomposition rate constants were significantly affected by temperature in the unfiltered RAS water sample (Fig. 1). With values of 0.979 h−1 at 17 degrees-C and 1.425 h−1 at 22 degrees-C, a temperature coefficient of 1.078 (7.8 percent increase per degree-C) was calculated.
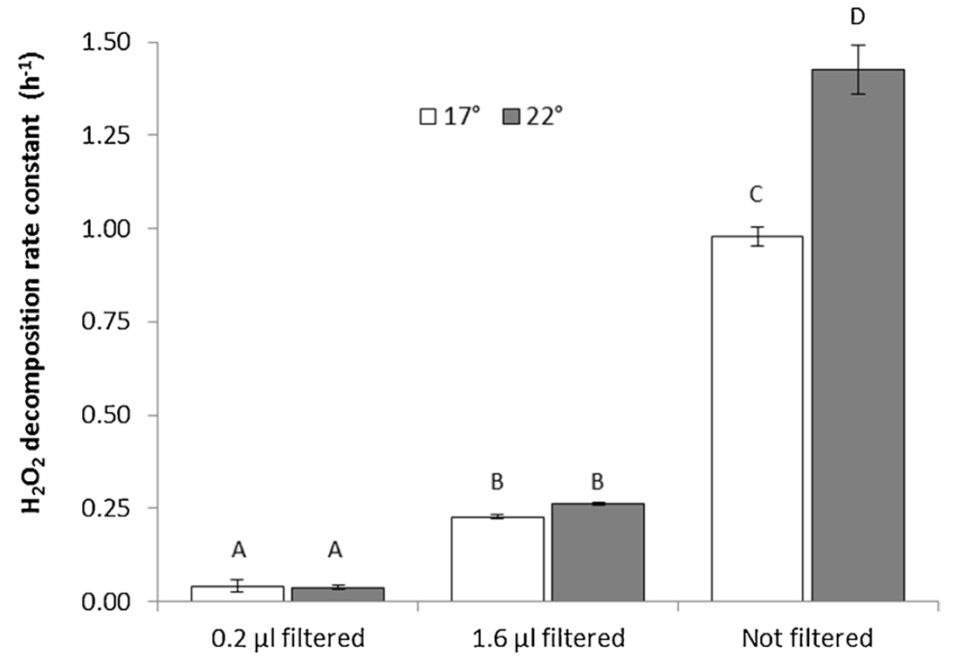
The H2O2 decomposition rate constants reached 3.7/hours at 15 degrees-C. As BOD5 reflects the amount of bioavailable dissolved and particulate organic matter, this parameter gives a more exact description of the bacterial growth potential than COD, which is nevertheless often used as a proxy due to its faster processing time. The H2O2 decomposition assay described here has potential to become a supplementary or alternative method to BOD5 and COD measurements since it correlates well with both parameters.
The H2O2 assay has a high reproducibility and allows fast measurements in both fresh and saline water samples where dilutions are often needed for BOD5 and COD. The H2O2 decomposition rate constant was also strongly correlated with the quantity of micro particles in RAS water samples. This means that our simple methods can be used as a proxy for both BOD5 and micro particle concentration.
The H2O2 decomposition assay has potential important applications. Bacteria in the water are suspended, aggregated and attached to particles, and they potentially challenge the performance of fish and RAS. The factors affecting microbial composition and dynamic, their virulence and associated causal relationships are likely to become a research area of high importance for the aquaculture industry in the near future.
Perspectives
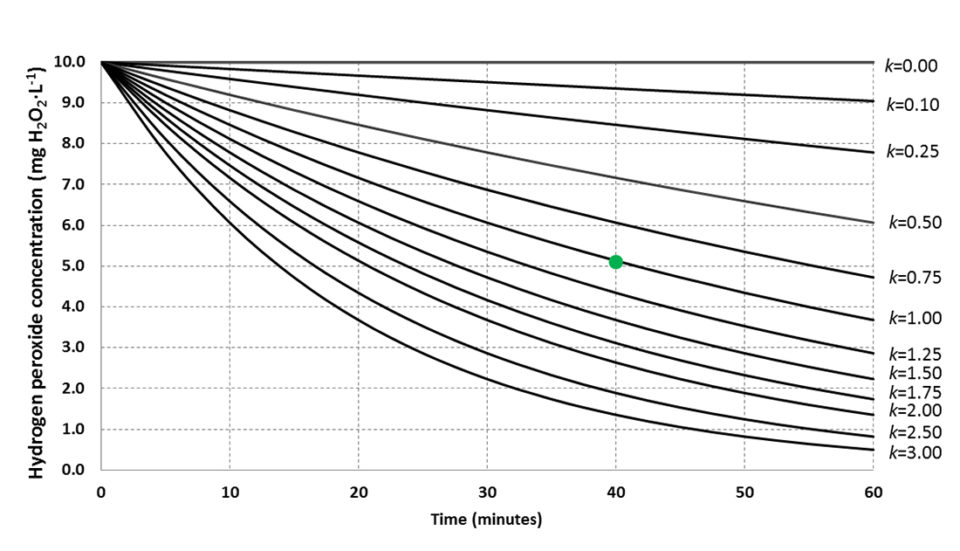
The assay presented here provides direct information on microbial activity in the water with a short-duration measurement. Fig. 2 conceptualizes how to apply the method in a practical way where an approximate rate constant is assessed based on time-flexible single measurements. In oligotrophic water, or water from advanced RAS with UV, ozone disinfection and efficient solids removal, increasing exposure time will improve resolution of the assay.
At the other end of the spectrum, the H2O2 assay can be made with shorter duration and increased H2O2 addition when assessing microbial rich eutrophic water from natural water bodies or open aquaculture pond systems with high feed loading and long water retention time.
As a fast and new tool to describe microbial activity, H2O2 decomposition assay are applicable for:
- Evaluating microbial activity in an aquaculture facility on a regular basis. This will give new information, increase system understanding and make it possible to establish baseline conditions and detect deviations.
- Quantifying and evaluating the effect of a given treatment component (foam fractionator, mechanical and biological filters etc.) and changes in operational practices (disinfection, new feed, altered fish densities, hydraulic, etc.).
- Improving planning and optimizing water treatment and sanitation (disinfection demand).
- Providing measurements of microbial activity and dynamics in field and laboratory trials.
- Replacing COD (and BOD5) measurements particularly for saline samples.
The assay could also be modified to quantify bacterial activity in biofilms or on system level. Modifications of the method might include simplifications (strip sticks), automation (e.g. using an online H2O2 sensor), use of other reagents, or use of plate reader assays to increase throughput.
In conclusion, the study has demonstrated that hydrogen peroxide decomposition is a rapid, efficacious and feasible indicator of microbial activity in water samples. The H2O2 assay has several potential applications in aquaculture worldwide from pond farming to intensive RAS as it provides new knowledge of the microbial water quality conditions in a fast, easy, competitive and reliable way. The new method we describe here can most likely also be applied in other areas, for example waste water quality determination.
Further details can be found in the original publication: Pedersen, L.F., Rojas-Tirado, P.A., Arvin, E., & Pedersen, P.B. (2019). Assessment of microbial activity in water based on hydrogen peroxide decomposition rates.
Aquacultural Engineering, 85, 9-14.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-

Lars-Flemming Pedersen, Ph.D.
Senior Research Scientist
Technical University of Denmark
DTU Aqua, Section for Aquaculture
The North Sea Research Centre
P.O. Box 101, DK-9850 Hirtshals, Denmark[107,100,46,117,116,100,46,97,117,113,97,64,112,102,108]
-

Paula Rojas-Tirado, Ph.D.
Researcher
Norwegian Institute for Water Research, NIVA
Section for Aquaculture
Thormøhlensgate 53D, 5006 Bergen, Norway -

Prof. Erik Arvin
Professor Emeritus
Technical University of Denmark
Department of Environmental Engineering
DTU Environment, Bygningstorvet
Build. 115, DK-2800 Kgs. Lyngby, Denmark -

Per Bovbjerg Pedersen, M.Sc.
Head of Section
Technical University of Denmark
DTU Aqua, Section for Aquaculture
The North Sea Research Centre
P.O. Box 101, DK-9850 Hirtshals, Denmark

