Marine Biological Laboratory develops quantitative real-time molecular assay

Atlantic cod (Gadus morhua) continues to be an attractive species for farming in the northeastern United States and other areas worldwide. High consumer demand and declining wild stocks have contributed to the interest. Improved hatchery techniques and engineering advancements in grow-out technology, including net pens and open water ranching, have also facilitated the developing cod-farming industry.
However, the uncertainties and high costs associated with raising cod fingerlings are still concerns for potential cod farmers in North America. One threat to the industry is nodavirus, which has demonstrated the capability of wiping out an entire season’s production of hatchery-sized cod.
Nodaviruses
Nodaviruses are a family of small viruses that infect the central nervous systems of fish. Numerous strains have been isolated from both wild and farmed fish.
The genomes of these viruses are composed of two single-stranded, positive-sense RNA molecules: RNA1 and RNA2. RNA1 is a putative RNA-dependent RNA polymerase, whereas RNA2 encodes for the capsid or coat protein.
Nodavirus infections cause viral encephalopathy and retinopathy, a condition that is also sometimes known as viral nervous necrosis. Symptoms of the disease include abnormal swimming behavior, swelling of the swim bladder, and altered skin coloration. The anorexia that has also been observed in infected cod could be associated with swim bladder pressure on the stomach and intestines. Mortality rates can be as high as 100 percent when hatchery populations are exposed to nodavirus.
New assay for screening
To assist hatcheries with broodstock screening and monitoring for nodavirus in fingerlings, the Marine Biological Laboratory in Woods Hole, Massachusetts, USA, developed a quantitative real-time molecular assay that recognizes the RNA2 gene of the Atlantic cod nodavirus. The research was supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, awarded to the Northeastern Regional Aquaculture Center at the University of Massachusetts Dartmouth.
In a single step, reverse transcription (RT) and polymerase chain reactions (PCR) are carried out on extracted RNA test samples using a fluorescence detection system. The assay monitors in actual time the fluorescence emitted during the PCR reaction as an indicator of nodavirus RNA2 presence. The RNA2 amplicon, or “piece” of RNA synthesized using PCR amplification techniques, is produced by a pair of unlabeled primers. The reaction also contains a dual-labeled oligonucleotide probe with 25 or fewer nucleotides and a melting temperature 10 degrees-C higher than the adjacent primers. The probe contains both fluorescent and quenching dyes.
When irradiated, the excited fluorescent dye transfers energy to the nearby quenching dye molecule rather than fluorescing. Close proximity of the reporter and quencher prevents emission of any fluorescence while the probe is intact.
The probe is designed to anneal to or align with a specific internal region of RNA2 in the area between where the pair of unlabeled primers anneal. When the polymerase replicates a template on which a probe is bound, its enzymatic activity cleaves the probe.
This ends the activity of the quencher and the reporter dye starts to emit fluorescence that increases in each cycle proportionally to the rate of probe cleavage. Accumulation of the amplicon is then detected automatically by monitoring the increase in fluorescence of the reporter dye.
The advantages of this dual-labeled probe nodavirus assay over conventional RT-PCR techniques include the fact the RT and PCR reactions and detection are all done in one sample well and processed in one instrument. Thus, multiple reaction wells and the separation of PCR products on agarose gels for detection are not required, saving time and resources, and eliminating added potential sources of error as samples are transferred and processed.
Successful testing
Using the dual-labeled RT-PCR assay, the author has detected nodavirus in the brain tissue of infected Atlantic cod. As expected, recently moribund fish had quantitatively higher levels of nodavirus compared to infected fish with minimal symptoms. Nodavirus has also been detected in blood samples from infected cod.
The assay has also been used to detect nodavirus in water samples from tanks holding infected fish, supporting the idea that horizontal transmission is an important vector of the disease. Detection in the water also raised the possibility that nodavirus can be brought into hatcheries from water sources.
This is the first time nodavirus was detected in both water and blood samples. A likely explanation is that the dual-labeled assay is far more sensitive than conventional molecular assays. For instance, a side-by-side comparison of serially dilute RNA extracted from the nervous tissue of a nodavirus-infected cod found the dual-labeled probe assay could detect over four orders of magnitude less nodavirus than conventional RT-PCR.
The detection in blood suggests that a nonlethal detection system could be developed to screen potential broodstock, as opposed to the current practice of removing nervous tissue following spawning events. Currently, however, the presence of nodavirus in blood is not consistently detected in fish found positive when nervous tissue is assayed. Further research is required to understand the complete etiology of Atlantic cod nodavirus and develop additional strategies to manage the problem.
(Editor’s Note: This article was originally published in the April 2005 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Steven Roberts, Ph.D.
Marine Biological Laboratory
7 MBL Street
Woods Hole, Massachusetts 02543 USA[117,100,101,46,108,98,109,64,115,116,114,101,98,111,114,115]
Tagged With
Related Posts

Responsibility
A helping hand to lend: UK aquaculture seeks to broaden its horizons
Aquaculture is an essential contributor to the world food security challenge, and every stakeholder has a role to play in the sector’s evolution, delegates were told at the recent Aquaculture’s Global Outlook: Embracing Internationality seminar in Edinburgh, Scotland.
Aquafeeds
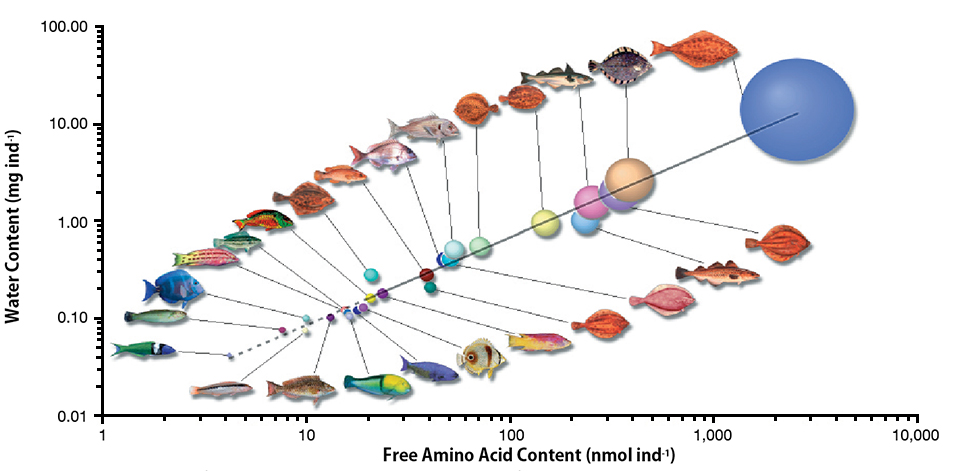
Amino acid requirements in developing marine fish
Most marine fish have pelagic eggs with large pools of free amino acids of fairly constant proportion regardless of species. The amino acids provide energy for metabolism and are an important nutritional asset for growing embryos.

Innovation & Investment
Aquaculture Exchange: Dawn Purchase, part 2
The Marine Conservation Society’s aquaculture program manager discusses what consumers ultimately need to know about aquaculture, responsible production and eco-labels. She also looks at NGOs making a difference in the global seafood industry and where she finds innovation in action.

Health & Welfare
Perspectives for red-banded sea bream culture
Red-banded sea bream could be farmed in systems designed for other sparid species. The authors’ facilities have achieved spontaneous spawning.


