Simulated commercial grow-out reinforces value for screening-based selection of IHHNV-free/low broodstock, use of SPF stocks and selection for IHHNV resistance/tolerance
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) was first identified in 1983 in the Americas as the cause of mass mortalities of farmed Pacific blue shrimp (Litopenaeus stylirostris). Soon thereafter it was also identified to cause shell deformities and stunted growth known as “runt deformity syndrome” in farmed Pacific white shrimp (Litopenaeus vannamei).
In Australia and elsewhere in the Indo-Pacific, IHHNV is commonly detected at high prevalence in wild and farmed black tiger shrimp (Penaeus monodon). While IHHNV infection has been considered to be relatively benign and of little consequence to the aquaculture of this species, acute infection was inferred to be the cause of severe shell deformities in a cohort of domesticated Generation 3 P. monodon reared in Indonesia in the early 1990s.
PCR-based detection of IHHNV has been complicated by genome sequence variation among strains of its three known lineages (I, II and III) and by non-infectious IHHNV genome forms having become integrated in the chromosomal DNA of some P. monodon within populations dispersed widely across its natural distribution range.
To avoid the cross-detection of integrated IHHNV genome forms, conventional and real-time PCR tests have been designed to either exclude their amplification or specifically detect the integrated IHHNV endogenous viral element at the exclusion of the infections IHHNV lineages. The real-time quantitative (q)PCR tests for IHHNV also provides a means of accurately quantifying IHHNV DNA amounts as a measure of relative infection loads.
This article summarizes the results of a study (https://doi.org/10.1016/j.aquaculture.2018.09.032) which was a grow-out trial of two cohorts of P. monodon that differed in their IHHNV prevalence and loads resulting from IHHNV infections loads differing substantially among the female broodstock from which they were derived. The shrimp were reared under simulated commercial conditions in four 0.16-hectare research ponds. Real-time qPCR was used to track IHHNV load and prevalence among groups of 48 shrimp sampled at regular intervals over the grow-out from each of the four ponds. These data identified a clear association between the early onset of high-level IHHNV infection and substantially reduced growth rates, survival and harvest yields.
This research was supported financially by the Fisheries Research and Development Corporation (FRDC), Australian Prawn Farmers Association (APFA), the Science and Industry Endowment Fund (SIEF) and CSIRO and is detailed in FRDC Project (2015-240 APFA IPA) Final Report ISBN 978–0–646-98,999-0. We thank the many individuals who supported the project in different ways and Ridley Aqua Feed for contributing to shrimp feed costs.
Grow-out trial setup
Penaeus monodon broodstock caught in coastal waters near Bramston Beach in North Queensland were transported by air to the Bribie Island Research Centre within 48 hours of capture. Upon receipt, each shrimp was sexed, weighed, eye-tagged and had a piece of pleopod tissue preserved for PCR analysis. Each of 2 x 10,000-liter circular maturation tanks were stocked with 32 females and 32 males. Tanks were housed in a darkened room, with light reducing lids, employed a 3-mm sand bed and were supplied heated flow-through seawater and aeration sufficient to maintain optimal dissolved oxygen levels. Broodstock were fed various natural feeds typical of those used in local commercial hatcheries to promote fecundity. An additional pleopod tissue was sampled from each female at the times when it was eyestalk ablated and spawned.
Eggs spawned from females were reared in tanks to postlarval stage 20 (PL20) using standard hatchery procedures. Pools of PL20 originating from either a group of three females or a group of four females were each stocked into two replicate plastic-lined and bird net covered grow-out ponds (40 m × 40 m × 2 m deep; 0.16 ha) and reared using standard local commercial practices. Groups of 144 shrimp from each of the four ponds were sampled at regular intervals over grow-out to record weights and sex and to preserve pleopod and/or lymphoid organ (LO) tissue for subsequent PCR analysis. Shrimp were harvested by netting and ultimately pond draining between 150 to 170 days of culture (DOC) to determine final pond yields and shrimp survival rates.

Please refer to Sellers et al. (2019) (https://doi.org/10.1016/j.aquaculture.2018.09.032) for more detailed information on the broodstock maturation and spawning methods, larval rearing, PL counting and pond stocking methods, shrimp grow-out and sampling methods as well as TNA extraction, cDNA synthesis and TaqMan real-time qPCR testing and data statistical analyses.
Pond yield and survival differences
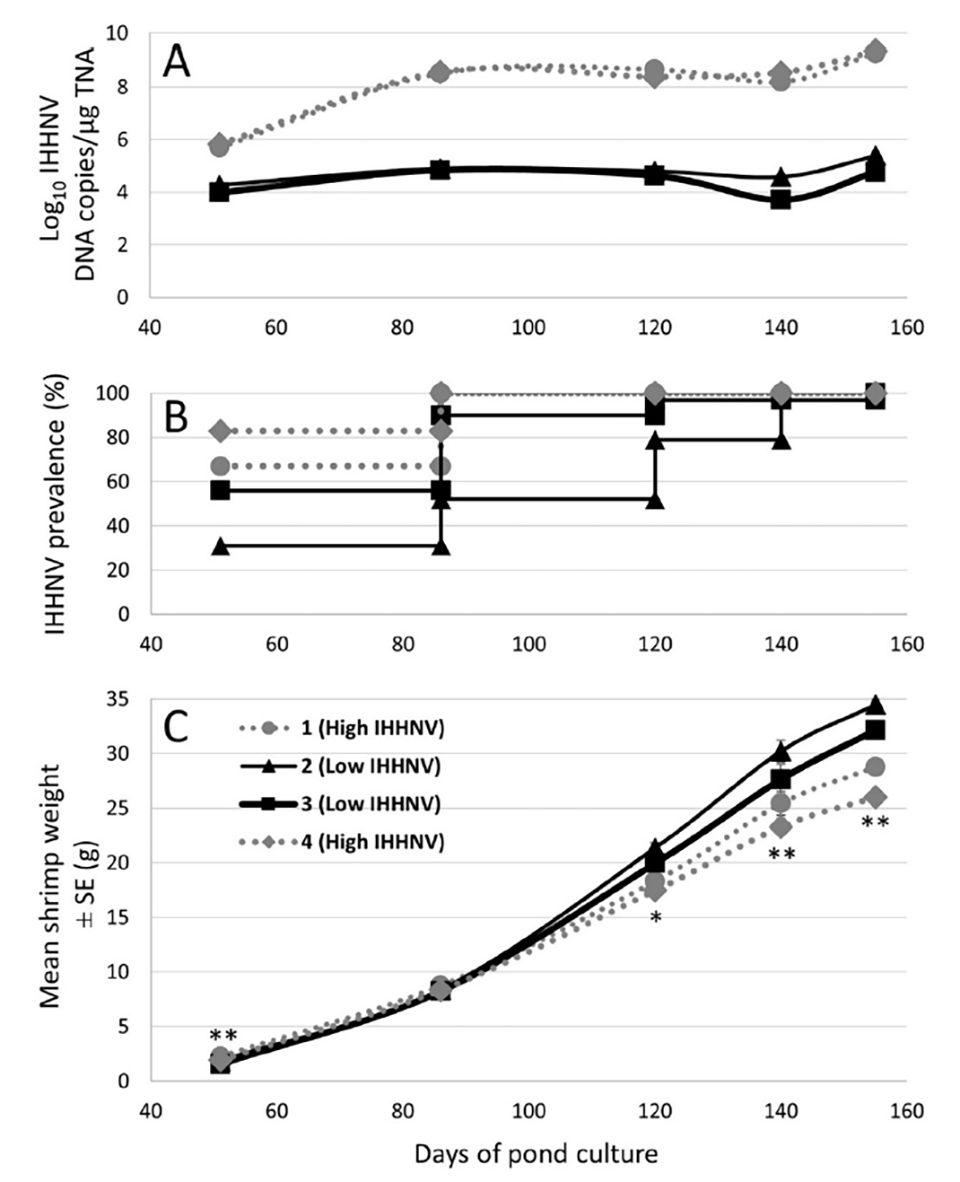
Weights of 144 P. monodon collected at random from each of the 4 x 0.16 ha research ponds were tracked at regular intervals over the grow-out. From 120 days of culture (DOC) onwards, weights were significantly lower among shrimp from the cohort stocked into 2 of the 4 ponds (ie. ponds 1 and 4; Fig. 1). The general appearance of shrimp collected from these two ponds from this time point onwards was also discernably reduced compared with those collected from ponds 2 and 3.
Shrimp were harvested progressively over a two-week period starting at 155 DOC. To quantify differences at a commercial scale, the final harvest yield realized in each 0.16-ha research pond was determined and extrapolated (x 6.25) to a typical 1-ha commercial pond. Using these data, yields from Ponds 2 and 3 (12.3 and 14.2 tons, respectively) were 22 percent to 58 percent higher than yields from ponds 1 and 4 (9.0 and 10.1 tons, respectively) (Table 1). Based on the average weights determined for the harvested shrimp, estimated survival in ponds 2 and 3 (95.9 percent and 99.8 percent, respectively) was also 13 percent to 25 percent higher than in ponds 1 and 4 (79.9 percent and 84.5 percent, respectively). As a result of the higher growth rate and survival of the shrimp cohort reared in ponds 2 and 3, collectively they consumed 36 percent more feed than the cohort reared in ponds 1 and 4.
Sellars, IHHNV, Table 1
| Pond No. | IHHNV Group | Feed consumed (kg) | Harvest weight (kg) | Feed conversion ratio (FCR) | Yield (MT/ha*) | Survival (%) |
|---|---|---|---|---|---|---|
| 2 | IHHNV-low | 3,245 | 2,273 | 1.43 | 14.2 | 95.9 |
| 3 | IHHNV-low | 3,337 | 1,996 | 1.70 | 12.3 | 99.8 |
| 1 | IHHNV-high | 2,434 | 1,613 | 1.51 | 10.1 | 84.5 |
| 4 | IHHNV-high | 2,388 | 1,436 | 1.66 | 9.0 | 79.9 |
* extrapolated from the 0.16 ha pond (i.e. total weight/0.16)
Using the extrapolated numbers, ponds 2 and 3 collectively produced 7.44 tons (av = 3.72 tons/ha) more shrimp that ponds 1 and 4. As these shrimp had a mean harvest weight resulting in 35 to 38 pieces/kg with a typical cooked wholesale value in Australia of about $18 AUD/kg, this 3.72 ton/ha additional yield would improve gross value of the crop (not considering additional feed, cooking and ancillary costs) by about $67,000 AUD per 1-ha pond.
IHHNV infection load differences
TaqMan real-time qPCR testing identified the absence of either gill-associated virus (GAV; yellow head virus genotype 2 (YHV2)) or yellow head virus genotype 7 (YHV7) in the broodstock and their progeny cohorts reared in the four ponds. Similar testing was thus undertaken to determine whether IHHNV infection might have been the cause of the reduced growth performance of the shrimp cohort reared in ponds 1 and 4.
Normalized amounts of TNA were tested by TaqMan real-time quantitative (q)PCR to directly compare IHHNV DNA loads (as a de facto measure of infection load/severity) across samples. qPCR data on pleopod tissue sampled at the time broodstock were received from North Queensland identified extremely low IHHNV loads in five and substantially higher loads in one female from each group contributing progeny stocked into either ponds 1 and 4 or ponds 2 and 3. The highest load (6.71 x 105 IHHNV DNA copies/µg TNA) was detected in 1 of the 3 females that contributed progeny to ponds 1 and 4.
IHHNV loads generally increased over the ~6 week period broodstock were matured, eyestalk ablated and spawned. However, loads increased to the highest levels in the three females contributing pond 1 and 4 progeny, with the highest female over 100-fold higher than the highest of the four females contributing pond 2 and 3 progeny. Testing of pools of eggs identified IHHNV loads to be highest, and >100-fold higher than eggs from any other female, in the pool collected from the female identified to possess the highest IHHNV loads when sampled both upon arrival and at the time it spawned.
Real-time qPCR was also used to detect and quantify IHHNV loads in pleopods from 48 of the 142 to 144 individual shrimp collected and weighed from each of the four ponds at 51, 86, 120 and 155 DOC as well as from 30 shrimp sampled from each pond at 140 DOC (Fig. 1). Among the shrimp tested at these times from ponds 1 and 4, IHHNV was detect at 100 percent prevalence from 86 DOC onwards. In contrast, among the shrimp tested from ponds 2 and 3 at the same times, IHHNV prevalence was lower at the earlier culture times and only reached 100 percent at sampling times (140-155 DOC) very late in the grow-out. Mean IHHNV loads were in the order of 100-fold higher in ponds 1 and 4 compared to ponds 2 and 3 at the initial sampling time point (51 DOC), and over 1000-fold higher at the four later sampling time points (Fig. 1).
Conclusions and implications
IHHNV infection has typically been witnessed to be relatively benign in aquaculture production of black tiger shrimp (Penaeus monodon). Despite this general premise, described here are findings showing that IHHNV infection in this species needs to be considered more seriously than previously thought. Under circumstances in which shrimp were encumbered by a sustained very high-load infection burden, IHHNV was found to be capable of profoundly compromising growth, general health, survival and pond harvest yields. With the quantum of yield losses estimated to be worth $67,000 AUD gross for a 1-ha commercial pond, measures that prevent such infections from arising are worthy of considering.
In regard to what interventions might offer the most effective means of restricting high-load infections occurring, it was noted that one of the seven female bloodstock used that had the highest pre-existing IHHNV loads upon capture from the wild also had the highest loads at the times it was eyestalk ablated and spawned.
Moreover, eggs produced from this female possessed IHHNV at loads (2.3 x 105 IHHNV DNA copies/µg TNA) massively higher than any of the other six females (30 to 1030 IHHNV DNA copies/µg TNA), irrespective of the fact that IHHNV loads had also increased moderately to massively in these females over the six-week period between when they were received and spawned. Thus, in cases when IHHNV is highly prevalent in wild broodstock captured for use in hatcheries, and capabilities exist to quantify infection severity by qPCR, such testing should be used as a means of identifying and culling such high-load females prior to them being conditioned and spawned. The qPCR screening of egg pools to identify and cull any with high loads of IHHNV should also be considered in such circumstances to ensure that only IHHNV-free or IHHNV-low seedstock are farmed.
Opportunities exist for RNA interference (RNAi) strategies to reduce loads of preexisting viral infections and thus the propensity for infections to be transmitted vertically to progeny. However, this technology has yet to mature to the point of providing a near-term solution, except possibly for assisting in clearing low-load infections from domesticated breeding lines of P. monodon. Such specific pathogen free (SPF) breeding lines of P. monodon would provide the solution, with lines also selected for enhanced IHHNV resistance/tolerance providing an even more ideal solution.
However, in countries like Australia where quarantine regulations strictly ban the import of live shrimp for aquaculture purposes, efforts to generate and perpetuate such breeding lines have not been able to break the cycle of reliance on wild-captured broodstock. Despite some progress, until such SPF lines can be established or acquired, screening-based selection will remain the only means of precluding IHHNV from causing production impacts on P. monodon farmed in Australia and elsewhere with the same predicaments.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aquaculture.2018.09.032.
References available from first author or original publication.
Authors
-

Melony J. Sellars, Ph.D.
Corresponding author
Queensland Bioscience Precinct
306 Carmody Road
St. Lucia, QLD 4067, Australia[32,117,97,46,111,114,105,115,99,64,115,114,97,108,108,101,83,46,121,110,111,108,101,77]
-
Jeff A. Cowley, Ph.D.
Queensland Bioscience Precinct
306 Carmody Road
St. Lucia, QLD 4067, Australia -
Dean Musson
Bribie Island Research Centre
144 North Street
Woorim, QLD 4507, Australia -
Min Rao
Queensland Bioscience Precinct
306 Carmody Road
St. Lucia, QLD 4067, Australia -
M.L. Menzies
Queensland Bioscience Precinct
306 Carmody Road
St. Lucia, QLD 4067, Australia -
Greg J. Coman, Ph.D.
Bribie Island Research Centre
144 North Street
Woorim, QLD 4507, Australia -
Brian S. Murphy
Bribie Island Research Centre
144 North Street
Woorim, QLD 4507, Australia[117,97,46,111,114,105,115,99,64,121,104,112,114,117,77,46,110,97,105,114,66]
Tagged With
Related Posts

Health & Welfare
Measuring susceptibility differences to IHHNV infection
This study evaluated the susceptibility to IHHNV in three different shrimp batches by measuring infectivity titers. Each batch showed a different infectivity titer, therefore, each batch had a specific susceptibility to IHHNV.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
EHP a risk factor for other shrimp diseases
Laboratory challenges and a case-control study were used to determine the effects of EHP infection on two Vibrio diseases: acute hepatopancreatic necrosis disease (AHPND) and septic hepatopancreatic necrosis (SHPN).

Health & Welfare
Four AHPND strains identified on Latin American shrimp farms
Two virulence genes are known to encode a binary photorhabdus insect-related toxin that causes acute hepatopancreatic necrosis disease in shrimp. The pathogenicities of these V. campbellii strains were evaluated through laboratory infection and subsequent histological examination in P. vannamei shrimp.


