Results to date of an applied selective breeding program in Egypt

The Indian white shrimp (Fenneropenaeus indicus) is an important species in the aquaculture industry of Egypt, and it has been negatively affected by various major diseases. Several selective breeding programs for farmed shrimp have succeeded elsewhere in improving disease resistance against specific pathogens, indicating that genetic improvement in resistance to several diseases is feasible.
However, for optimal implementation of selection for improved disease resistance in a breeding program, it is critical to investigate if significant genetic correlations exist between disease resistance and other traits of importance included in the breeding objective.
The selection for disease resistance is a sustainable method for developing aquaculture production by offering ways to develop more disease resistant stocks. It is well documented that there is large additive genetic variance in resistance to common diseases in aquaculture, and this may facilitate high response to selection. Efficient breeding programs for shrimp should include improved disease resistance through selection.
Our research work implemented selection for increased disease resistance as a part of the selection program for Indian white shrimp at a private shrimp farm in the AL Dibah Triangle Zone (DTZ) in Egypt. Our research is based on controlled challenge test experiments in which shrimp from each of the individual families produced in the breeding nucleus are exposed to WSSV, IHHNV and vibrio (EMS) using infected cohabitants (disease carriers).
The overall objectives of our project – supported by Science & Technology Development Fund (STDF), Egypt under grant No. 5661 – were to develop a selection program for increased resistance to major shrimp pathogens in Indian white shrimp, to investigate the level of additive genetic variation for resistance to a combined infection of WSSV, IHHNV and vibrio (EMS) in challenge tests under commercial rearing conditions, and to develop a single, all-purpose, strain or breed rather than several breeds specialized for particular environments (e.g. particular diseases).
We hope our work has direct economic benefits for shrimp farmers, by reducing shrimp losses due to diseases outbreaks. Here we review some of the project’s main activities since late 2013.

Development of microsatellite markers
One of our objectives is to develop a genotyping system for Indian white shrimp (Fenneropenaeus indicus) resistant to various diseases, using microsatellite loci as markers. This system will eventually be used to genotype the broodstock of these species in order to monitor genetic diversity within this group as a form of broodstock management.
The markers and resulting genotype data will be used primarily for four purposes: (1) comparison of broodstock genetic diversity among shrimp populations and estimation of genetic divergence among populations (e.g. Fst), (2) monitoring secular changes in allele frequencies in closed broodstock, (3) parentage assignment and full-sib family identification in broodstock, (4) forensic and intellectual property applications.
In phase I, 100 primers were designed from microsatellite sequences of F. indicus. For the optimization and polymorphism testing of phase II, 41 primers were initially tested. Currently, 21 primer sets have been optimized and used to amplify microsatellites in the individuals in phase I. To date, 9 of these loci have proven polymorphic, will provide useful information and will subsequently be used in genotyping. Polymorphism in the remainder of the loci is yet undetermined as they are still being analyzed on polyacrylamide gels. If necessary, more loci will be added to the original 21 from the 41 primer sets that have been ordered. Completion of loci selection for use in genotyping is targeted for the end of 2016.
Specialized strains for environments
The original goal was to produce one, all-purpose, improved strain of Indian white shrimp, but we have evolved our goal and committed ourselves to two very different growout systems, intensive and semi-intensive. The new emphasis on intensive environments implies that many of the shrimp will grow their entire lives at high densities without natural food (except for biofloc) and, possibly, without some of the disease challenges found in ponds. However, we also remain committed to semi-intensive cultivation in environments where there are natural food, predators and space for behavioral interactions.
There may be trade-offs among traits that affect growth and survival in these two environments. Although I am not aware of any evidence of such trade-offs in shrimp – or fish for that matter – they are found in other domestic animals. The possibility of trade-offs between growth and disease resistance in either or both environments, remains open.
Therefore, the genetics program will develop only one strain for the first two generations and until January 2016. In the second half of this year, this information allowed us to make an informed decision about how many strains to develop from then on. The required information included growth and survival records on individuals which have known family relationships with each other, and is the same information used for selection. Collection of this information should be a routine component of any genetic programs.

Relative selection intensity on growth and survival
During the first year, we worked on equal selection intensities on growth and resistance to WSSV, IHHNV and vibrio (EMS) challenges, but more recently we adopted a relative weighting of 70:30, respectively. The balance will be achieved by adjusting the severity of growth selection and WSSV, IHHNV and vibrio (EMS) challenges, and by skipping the challenge entirely in some selected families or batches of families.
Decisions on whether or not to challenge any particular group of animals can easily be made on the fly by the project’s genetics manager if record-keeping is up to date. Analysis of possible trade-offs between growth and survival were made during the major review in 2015 when the selection program was revised.
Incorporating improved broodstock into the production system
The plan in early 2014 was to produce each “family” of offspring from four brothers mated with four sisters. The selection intensity on survival in 2015 was reduced and there is now increased emphasis on protecting the broodstock from piracy. For these reasons, our new approach is to have a family breeding system in which only one male and one female (not 4 of each) are used to produce each of the families within which selection takes place. A brother of the genetics program male and a sister of the female will be mated for the production line.
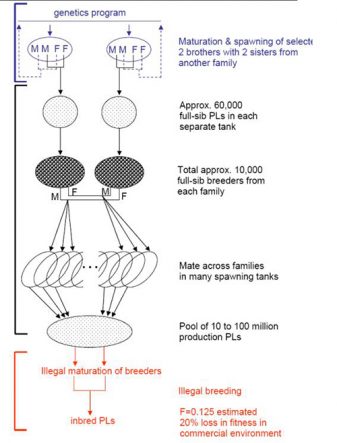
The downside of this decision is that the performance of the production shrimp postlarvae (PL) will always be one generation behind the performance of the selection line. Production performance will improve at the same rate as the selection line with a lag of one generation.
Genetic protection against piracy
Originally, we thought that keeping ahead of the competition by producing better PL every year would be sufficient protection for our work, at least until a microsatellite-based identification system could be put in place. Now it seems that an improved strain can be pilfered almost as quickly as we can develop it. Therefore, the production system has been modified so that any production PL that are used as breeders will produce offspring that are uniformly inbred at F=0.125, which is equivalent to half-sib mating.
The resulting inbreeding depression is around 20 percent under the stressful conditions of aquaculture production. Subsequent generations of the pirated broodstock will have an average F=0.125, and as an additional challenge, the inbreeding will be variable among individuals and batches, making growout performance rather unpredictable. This genetic protection against piracy is the principal reason why the farmed production of PL will lag one year behind the PL from the genetic program.
Evidence from microsatellite markers for farmed shrimp inbreeding and disease outbreaks in Egypt
The disease crisis faced by the shrimp farming industry in Egypt is a result of the interaction between management practices that cause inbreeding, the presence of disease pathogens, and the animals’ susceptibility to disease and environmental stresses. The inbreeding increase has promoted the incidence of various diseases, including WSSV, IHHNV, YHV, vibrio and EMS (AHPND).
We started a breeding program in 2013 for the selection of disease-resistant Indian white shrimp in Egypt as an applied commercial breeding program to improve the production and profitability in shrimp aquaculture in the country. At the time, only a portion of the population contributed genes transmitted to the next generation, reducing effective population sizes, and shrimp farmers were mating related shrimp and using inbred lines.
This increased the degree of inbreeding over generations, and this was noticed in the traits of interest, such as disease resistance, growth and reproductive performance. Therefore, in our breeding program for disease resistance and growth in Indian white shrimp, we are regularly monitor and quantifying the effects of inbreeding and proper balancing of the response to selection and the effects of inbreeding.
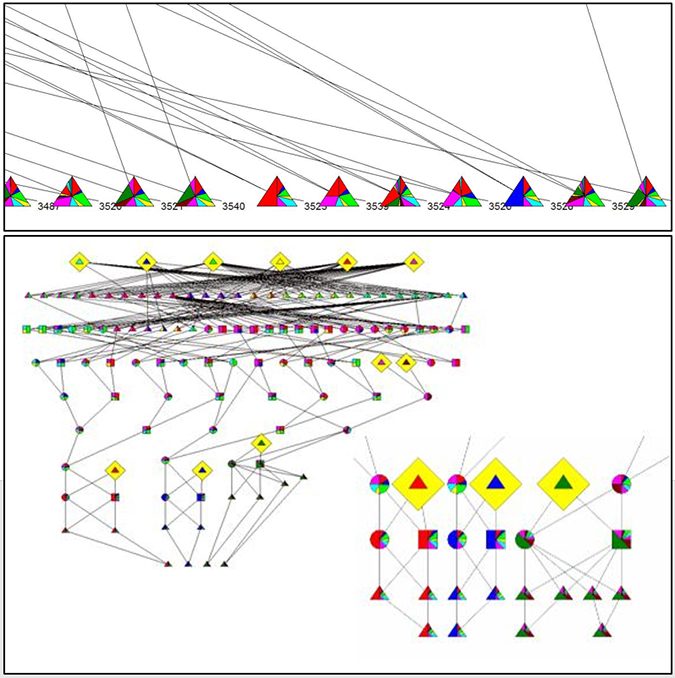
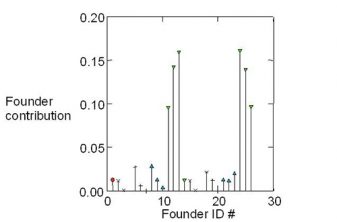
Microsatellite markers and pedigree were both used to assess genetic variability and effective population size. Using 10 loci, selected shrimp populations for three generations showed a decline in the expected heterozygosity (15 percent) and allelic diversity indices (52 to 93 percent), compared to the wild population (P < 0.05). Effective population size estimates based on microsatellites declined from 46.5 to 77.0 percent in cultured populations (P < 0.05), compared to the wild population. Working in direct contact with stakeholders has led to improvement in dissemination of the knowledge and awareness.
Our strategy to control long-term inbreeding in breeding program is to use a relatively large number of broodstock in each generation, and to control mating among relatives by imposing mating restrictions (use of microsatellite markers for paternity analysis and mate design). The results have shown that the use of microsatellite markers suggest that heterozygosity at microsatellites loci is well correlated with individual inbreeding coefficients. The decline of genetic variability in the cultured selected population due to domestication, and evidence of a further smaller decline in effective population size across generations in the selected population, were observed when analyzing pedigree and microsatellite data.
Pedigree keeping is required to prevent the decline of effective population size and maintain genetic variability in shrimp breeding programs, while microsatellites are useful to assess effective population size changes at the population level. However, we still allow preservation of the genetic variation necessary to continue improving the population for current traits or others that may need to be incorporated in the future.
Additional information and references are available from the author.
Author
-

Mohamed EL. Sayed Megahed, Ph.D.
Research Scientist
National Institute of Oceanography and Fisheries (NIOF)
Ministry of Scientific Research
Gulfs of Suez & Aqaba’s Branch
Suez
Egypt[109,111,99,46,108,105,97,109,103,64,101,118,111,114,112,109,105,101,103,97,117,113,97]
Tagged With
Related Posts

Health & Welfare
Shrimp biofloc production trials in Saudi Arabia
An R&D trial was conducted with shrimp biofloc technology at a commercial facility in Saudi Arabia. Results indicated that biofloc technology can be effectively applied, with some protocol adjustments, in the KSA’s extreme desert environmental conditions.

Responsibility
Genetic improvement aids red tilapia growth in Egypt
A new breeding program for genetic improvement of red tilapia was established at the Fish Research Center (FRC), Suez Canal University, in Ismailia, Egypt. It aims to improve the growth rate of the fish and to provide significant benefits to tilapia farmers.

Health & Welfare
Successful production of Nile and blue tilapia fry
The relationship between production in subtropical earthen ponds of fry suitable for hormonal sex inversion and degree days was quantified for Nile tilapia (Oreochromis niloticus; Egypt strain) and blue tilapia (O. aureus).

Intelligence
India a unique opportunity for rainbow trout farming
India has a small trout farming industry in the Himalayan and Southern Peninsula areas. Even though current annual production is only a few hundred tons, the country has many areas that, although geographically difficult to work in, also offer excellent, very large habitats and conditions for trout culture.

