In vitro method synthesizes and amplifies specific sequences of target DNA
The Polymerase Chain Reaction (PCR) is a very prominent molecular biology technique, originally conceived by Dr. Kary Mullis during a moonlit mountain drive in California in 1983. It dawned on him that the property of DNA to separate into single strands at warm temperatures and the ability of the polymerase enzyme to exactly replicate single strands could be used together to multiply sequences of DNA. Dr. Mullis received the Nobel Prize for chemistry in 1993 for his development.
PCR is a technique that is used to amplify the number of copies of a specific region of DNA, in order to produce enough DNA to be adequately tested. It is, essentially, an in vitro method that synthesizes and amplifies specific sequences of target DNA through a repeating enzymatic thermocycling process. In each cycle the amount of target DNA is doubled until there is enough to be detected by gel electrophoresis, another common molecular biology technique.
The development of PCR means that small amounts of DNA no longer limit molecular biology research or DNA-based diagnostic procedures. The technique is continuously improving and its full impact on molecular diagnostics is yet to come. PCR currently has many applications, including analysis of ancient DNA from fossils, amplification of small DNA amounts for analysis by DNA fingerprinting, mapping the human genome and also those of other species, and detection of microorganisms present in low densities in water, food, soil or other organisms. In aquaculture PCR is rapidly becoming a vital tool in the detection, management and prevention of viral diseases in penaeid shrimp culture worldwide.
PCR is used to produce copies of a specific region of DNA. This technique can be used to identify with a very high-probability, disease-causing viruses and/or bacteria, a deceased person, or a criminal suspect. In order to use PCR, one must already know sequences that flank both ends of a given region of interest in the DNA (may be a gene or any sequence). One need not know the DNA sequence inbetween. The building-block sequences (nucleotide sequences) of many of the genes and flanking regions of genes of many different organisms are known. While some genes may be the same, or very similar among organisms, there will always be genes with DNA sequences that differ among different organisms. Therefore, by identifying the genes that are different, and therefore unique, one can identify an organism.
A gene’s building-block sequence is the precise linear order of appearance of four different components (deoxyribonucleotides) within a stretch of DNA (deoxyribonucleic acid). The four components are: Adenine, Thymine, Cytosine and Guanine, abbreviated as: A, T, C and G, respectively (a four-letter alphabet). The linear arrangement of the letters of this alphabet generates a “sentence” (a gene sequence). The number of letters in the sentence may be relatively few, or relatively many, depending on the gene. If the sentence is 1000 letters-long, the sequence would be said to be one kilobase (1000 bases). DNA is double-stranded (except in some viruses), and the two strands pair with one another in a very precise way. Each letter in a strand will pair with only one kind of letter across from it in the opposing strand: A always pairs with T, and C always pairs with G across the two strands.
 Now, let’s say that the above sequences “flank” (are on either end of..) the gene, which includes a long stretch of letters designated as: ………….. These are known, absolutely identified, to be the sequence of letters, which only flank a particular region of a particular organism’s DNA, and no other organism’s DNA. This region would be a target sequence for PCR.
Now, let’s say that the above sequences “flank” (are on either end of..) the gene, which includes a long stretch of letters designated as: ………….. These are known, absolutely identified, to be the sequence of letters, which only flank a particular region of a particular organism’s DNA, and no other organism’s DNA. This region would be a target sequence for PCR.
The first step for PCR would be to synthesize the flanking sequences, known as”primers” of about 20 letters-long. This step is easily done, by adding one letter-at-a-time in the order desired to a machine called a DNA synthesizer. In this example, the primers we wish to make will be exactly the same as the flanking sequences shown above. We make ONE primer exactly like the lower left-hand sequence, and ONE primer exactly like the upper right-hand sequence, to generate:
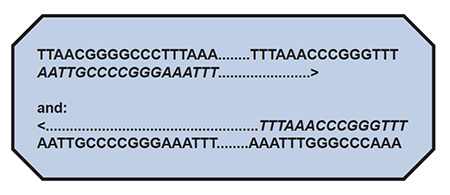 Now, the …….. may be a very long set of letters inbetween; it does not matter. If you look at this arrangement, you can see that if the lower left-hand primer sequence (italics) paired to the upper strand could be extended to the right in the direction of the arrow, and the upper right-hand sequence paired to the lower strand could be extended to the left in the direction of the arrow (remembering that the ……… also represent letters, and opposite pairing will ALWAYS be A to T and C to G), one could exactly duplicate the original gene’s entire sequence.
Now, the …….. may be a very long set of letters inbetween; it does not matter. If you look at this arrangement, you can see that if the lower left-hand primer sequence (italics) paired to the upper strand could be extended to the right in the direction of the arrow, and the upper right-hand sequence paired to the lower strand could be extended to the left in the direction of the arrow (remembering that the ……… also represent letters, and opposite pairing will ALWAYS be A to T and C to G), one could exactly duplicate the original gene’s entire sequence.
There would be four strands produced where originally there were only two. If the procedure is repeated, eight strands will be made, etc. About twenty cycles will produce approximately one million copies of the original sequences (2 raised to the 20th power). With this amplification potential, there is enough DNA in one-tenth of one-millionth of a liter (0.1 microliter) of human saliva (which contains shed epithelial cells), to use PCR to identify human DNA.
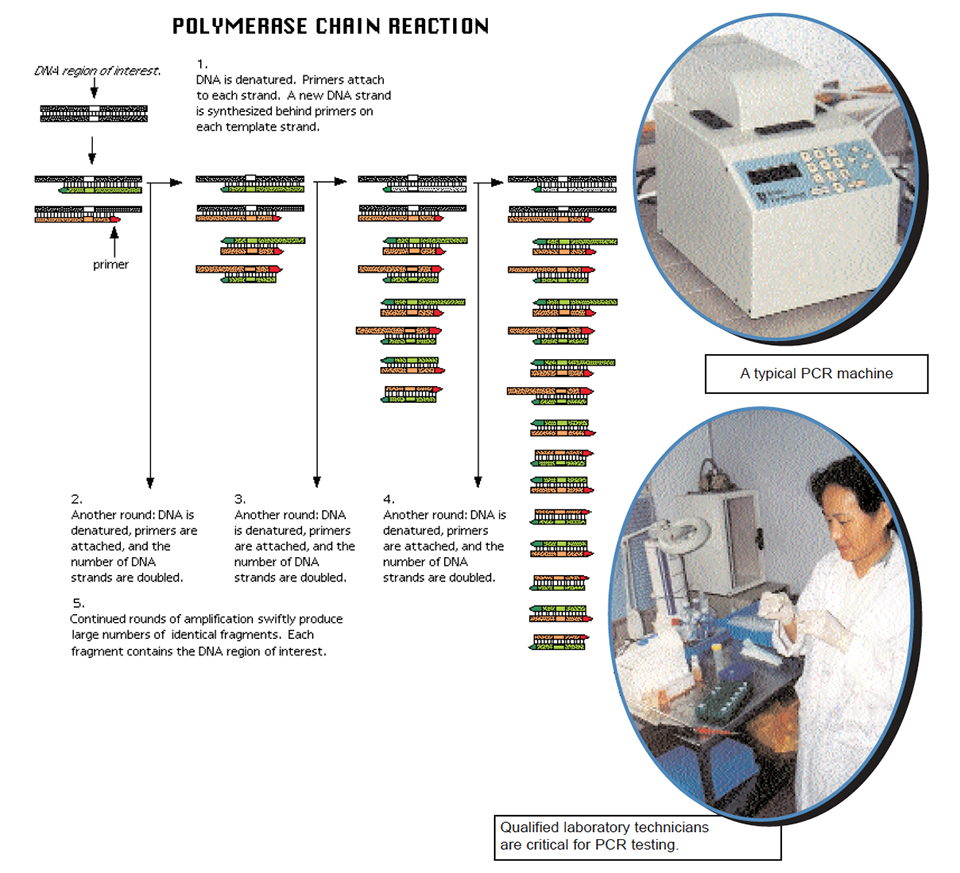
The following steps are generally used for PCR:
- Unknown DNA is heated, which causes the paired strands to separate (single strands now accessible to primers);
- Add large excess of primers relative to the amount of DNA being amplified, and cool the reaction mixture to allow double-strands to form again (because of the large excess of primers, the two strands will always bind to the primers, instead of with each other);
- To a mixture of all four individual letters (deoxyribonucleotides), add an enzyme that can “read” the opposing strand’s “sentence” and extend the primer’s “sentence” by “hooking” letters together in the order in which they pair across from one another – A:T and C:G. This particular enzyme is called a DNA polymerase (because makes DNA polymers). One such enzyme used in PCR is called Taq polymerase.
The enzyme synthesizes new DNA in opposite directions, but only this region of the entire DNA available. After one cycle, add more primers, add four-letter mixture, and repeat the cycle. The primers will bind to the “old” sequences as well as to the newly-synthesized sequences. The enzyme will again extend primer sentences. Eventually, there will be sufficient DNA copies of the region of interest with which to work. By use of specific primers that represent known flanking regions of different genes of various organisms in separate experiments, one can determine if DNA representative of a given organism is present or not. If no DNA is amplified, the primers did not bind to anything in the test sample. On the other hand, appearance of copies of a region of DNA by PCR allows precise identification of the source of amplified material.
PCR is a valuable tool for the prevention, control and management of various diseases in aquaculture. For shrimp farmers, it allows for widespread, fast and sensitive screening of potential shrimp virus carriers, and also for detection of early or light infections. The tests can be carried out non-destructively using body fragments, haemolymph or feces from shrimp tested. PCR can be used to screen broodstock animals and also postlarvae before stocking. PCR is a vital tool to properly detect White Spot and other shrimp pathogens, but unfortunately many shrimp producers still do not clearly understand the technique or its limitations. It can be an invaluable weapon against shrimp viruses, if properly used.
References
Dieffenbach, C.W & G.S. Dvksler. 1995. PCR primer: a laboratory manual. CSHL press, Cold Spring Harbor, USA.
Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. 1990. PCR Protocols: a guide to methods and applications. Academic Press, New York.
Ellingboe, J., & U. B. Gyllensten. 1992. The PCR technique: DNA sequencing. Eaton Publishing Co., Natick, MA.
Bej, A. K., M. H. Mahbubani, & R. M. Atlas. 1991. Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their applications. Critical Reviews in Biochemistry and Molecular Biology 26:301-334.
PCR Methods and Applications is a journal published quarterly by Cold Spring Harbor Press. Their address is: Cold Spring Harbor Laboratory Press, 10 Skyline Drive, Plainview, New York USA 11803-9729.
(Editor’s Note: This article was originally published in the August/October 1999 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
John C. Brown, Ph.D.
Professor and Director of Graduate Studies,
Department of Molecular Biosciences
University of Kansas
Lawrence, KS 66045
Tagged With
Related Posts

Health & Welfare
Effective biosecurity requires more than window dressing
Biosecurity in aquaculture involves actions designed to maintain the health of aquatic animals by reducing the introduction of pathogens to facilities.

Health & Welfare
The unmet promise of pondside PCR
A new generation of technology, portable PCR, offers potential for affordable, immediate, pondside diagnosis in easy-to-use handheld kits. But will it live up to the hype?
Health & Welfare
Rapid test detects NHP in penaeid shrimp
A rapid lateral-flow immunoassay using monoclonal antibody combinations can identify necrotizing hepatopancreatitis (NHP) infection in less than 15 minutes.
Health & Welfare
A rapid, sensitive RPA assay for detection of Vibrio parahaemolyticus in seafood
A real-time RPA method has been developed for the detection of Vibrio parahaemolyticus with high sensitivity and specificity.


